Note
Click here to download the full example code
Eighth Example: # Dynamically built component using input signal values during the fit stage¶
- We continue demonstrating several interesting features:
- How the user can choose to encapsulate several blocks into a PipeGraph and use it as a single unit in another PipeGraph
- How these components can be dynamically built on runtime depending on initialization parameters
- How these components can be dynamically built on runtime depending on input signal values during fit
- Using GridSearchCV to explore the best combination of hyperparameters
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.preprocessing import MinMaxScaler
from sklearn.mixture import GaussianMixture
from sklearn.linear_model import LinearRegression
from pipegraph.base import PipeGraph, RegressorsWithDataDependentNumberOfReplicas
X_first = pd.Series(np.random.rand(100,))
y_first = pd.Series(4 * X_first + 0.5*np.random.randn(100,))
X_second = pd.Series(np.random.rand(100,) + 3)
y_second = pd.Series(-4 * X_second + 0.5*np.random.randn(100,))
X_third = pd.Series(np.random.rand(100,) + 6)
y_third = pd.Series(2 * X_third + 0.5*np.random.randn(100,))
X = pd.concat([X_first, X_second, X_third], axis=0).to_frame()
y = pd.concat([y_first, y_second, y_third], axis=0).to_frame()
Now we consider the possibility of using the classifier’s output to automatically adjust the number of replicas. This can be seen as PipeGraph changing its inner topology to adapt its connections and steps to other components context. This morphing capability opens interesting possibilities to explore indeed.
import inspect
print(inspect.getsource(RegressorsWithDataDependentNumberOfReplicas))
Out:
class RegressorsWithDataDependentNumberOfReplicas(PipeGraph, RegressorMixin):
def __init__(self, steps=[('regressor', LinearRegression() )]):
self.steps = steps
def fit(self, *pargs, **kwargs):
selection = kwargs['selection']
selection = selection.iloc[:,0].values if isinstance(selection, pd.DataFrame) else selection
kwargs['selection'] = selection
regressor = self.named_steps.regressor
number_of_clusters = len(np.unique(selection))
multiple_regressors = RegressorsWithParametrizedNumberOfReplicas(number_of_replicas=number_of_clusters,
regressor=regressor)
steps = [('regressorsBundle', multiple_regressors)]
connections = dict(regressorsBundle={'X': 'X', 'y': 'y', 'selection': 'selection'})
self._pipegraph = PipeGraph(steps=steps, fit_connections=connections)
self._pipegraph.fit(*pargs, **kwargs)
return self
def predict_dict(self, *pargs, **kwargs):
return self._pipegraph.predict_dict(*pargs, **kwargs)
Using this new component we can build a simplified PipeGraph:
scaler = MinMaxScaler()
gaussian_mixture = GaussianMixture(n_components=3)
models = RegressorsWithDataDependentNumberOfReplicas()
steps = [('scaler', scaler),
('classifier', gaussian_mixture),
('models', models), ]
connections = {'scaler': {'X': 'X'},
'classifier': {'X': 'scaler'},
'models': {'X': 'scaler',
'y': 'y',
'selection': 'classifier'},
}
pgraph = PipeGraph(steps=steps, fit_connections=connections)
pgraph.fit(X, y)
y_pred = pgraph.predict(X)
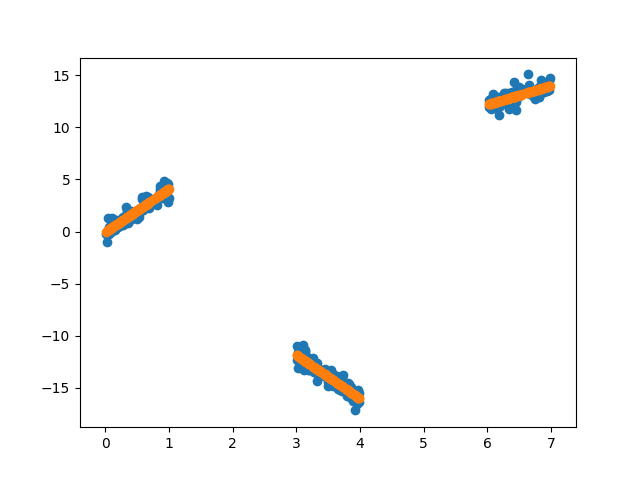
plt.scatter(X, y)
plt.scatter(X, y_pred)
Total running time of the script: ( 0 minutes 0.049 seconds)